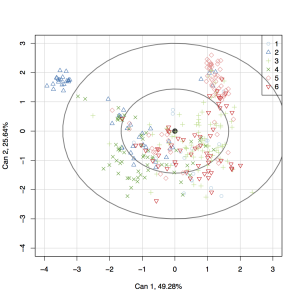
 Bayesian population genetics inference methods implemented in software packages such as Structure and InStruct are extensively used to infer the presence of population structure within genetic data. The primary outputs of these methods are ancestry profiles of individuals among a number of computationally inferred populations. Presently, these ancestry profiles are visualized using distruct, and there are no objective tests conducted to determine to what extent the factor of interest (usually geographic origin) correlates with inferred population structure, and if so, which populations are driving the structure.
Bayesian population genetics inference methods implemented in software packages such as Structure and InStruct are extensively used to infer the presence of population structure within genetic data. The primary outputs of these methods are ancestry profiles of individuals among a number of computationally inferred populations. Presently, these ancestry profiles are visualized using distruct, and there are no objective tests conducted to determine to what extent the factor of interest (usually geographic origin) correlates with inferred population structure, and if so, which populations are driving the structure.
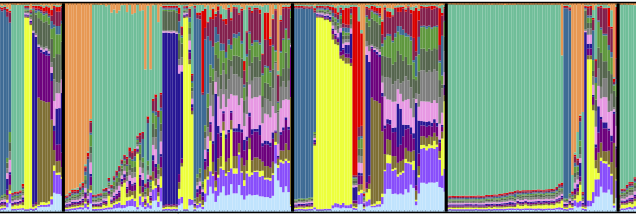 Here we present ObStruct, a novel application of a classic statistical method to objectively analyze these ancestry profiles to estimate the amount of structure in a data set, provide information on the relative contributions of sampled and inferred populations to this structure, test the statistical significance of differences between populations and crucially determine whether the factor of interest correlates with the inferred population structure. ObStruct works alongside existing analysis methods in population genetics and provides a useful final step in the pipeline for analyzing population genetics data.
Here we present ObStruct, a novel application of a classic statistical method to objectively analyze these ancestry profiles to estimate the amount of structure in a data set, provide information on the relative contributions of sampled and inferred populations to this structure, test the statistical significance of differences between populations and crucially determine whether the factor of interest correlates with the inferred population structure. ObStruct works alongside existing analysis methods in population genetics and provides a useful final step in the pipeline for analyzing population genetics data.
ObStruct Publication
You can view the entire ObStruct publication for free over at PLOS ONE.
Download ObStruct
ObStruct is available as a Perl script along with a manual describing its use and features.
Important note: If you are running Microsoft Windows you will need to install Perl in order to use ObStruct. We recommend Strawberry Perl which can be found here. If you’re running Apple OS X or Linux, relax, Perl already comes included with your operating system.
Enquiries and Support
If you have any enquiries about ObStruct, please email v.gayevskiy@auckland.ac.nz.
Licensing Information
![]() ObStruct is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported (CC BY-SA 3.0) License.
ObStruct is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported (CC BY-SA 3.0) License.